Evan Seitz
Research Scientist
InstaDeep
Cold Spring Harbor Laboratory
Simons Center for Quantitative Biology
Biography
I’m Evan Seitz, a computational biologist working at the intersection of machine learning, regulatory genomics, and structural biology. I recently joined InstaDeep as a Research Scientist, where I focus on advancing genomics using genomic large language models (gLMs) and other AI-driven approaches.
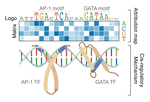
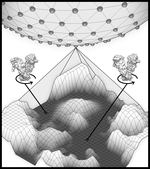
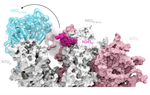
As a postdoctoral fellow at Cold Spring Harbor Laboratory with Drs. Peter Koo and Justin Kinney, I developed two explainable AI frameworks—SQUID and SEAM—for uncovering cis-regulatory mechanisms in genomic deep neural networks. Previously, I completed my Ph.D. with Nobel laureate Joachim Frank at Columbia University, where I developed geometric machine learning and explainability frameworks, including ESPER, to reveal conformational heterogeneity in cryo-EM protein structures.
My work is driven by a deep curiosity about nature, and how to leverage learning systems to extract meaning from biological complexity. I’m always open to conversations around research, collaboration, or industry roles at the intersection of AI and biology.
This site serves as a hub for my work, ideas, and ongoing projects in computational biology and machine learning. You can download my:
Or connect with me on LinkedIn.
- Machine Learning
- Explainable AI
- Gene Regulation
- Protein Conformational Heterogeneity
- Computational Biology
- Complex Systems
-
PhD in Biological Sciences, with Distinction (Geometric Machine Learning & Computational Biophysics)
Columbia University, 2017–2022
-
BS in Physics, with Highest Honor (Computational Biophysics)
Georgia Institute of Technology, 2015–2017
-
BA in Mass Communication
Georgia College, 2005–2009